There’s an enhancement to the GEDmatch AutoSegment clustering on Genetic Affairs. Now the GEDmatch option includes using the triangulated data as well as the all segment data, both of which are available on Tier 1 of GEDmatch.
There are a number of settings for the GEDmatch DNA Segment Search. I used 1000 for my analysis. Most of the settings can be left to the defaults. However, if you’re including matches that have long segments with you, you’d want to click the ‘Prevent Hard Breaks’ option. GEDmatch default adds in hard breaks when it finds segments over 500,000 base positions.
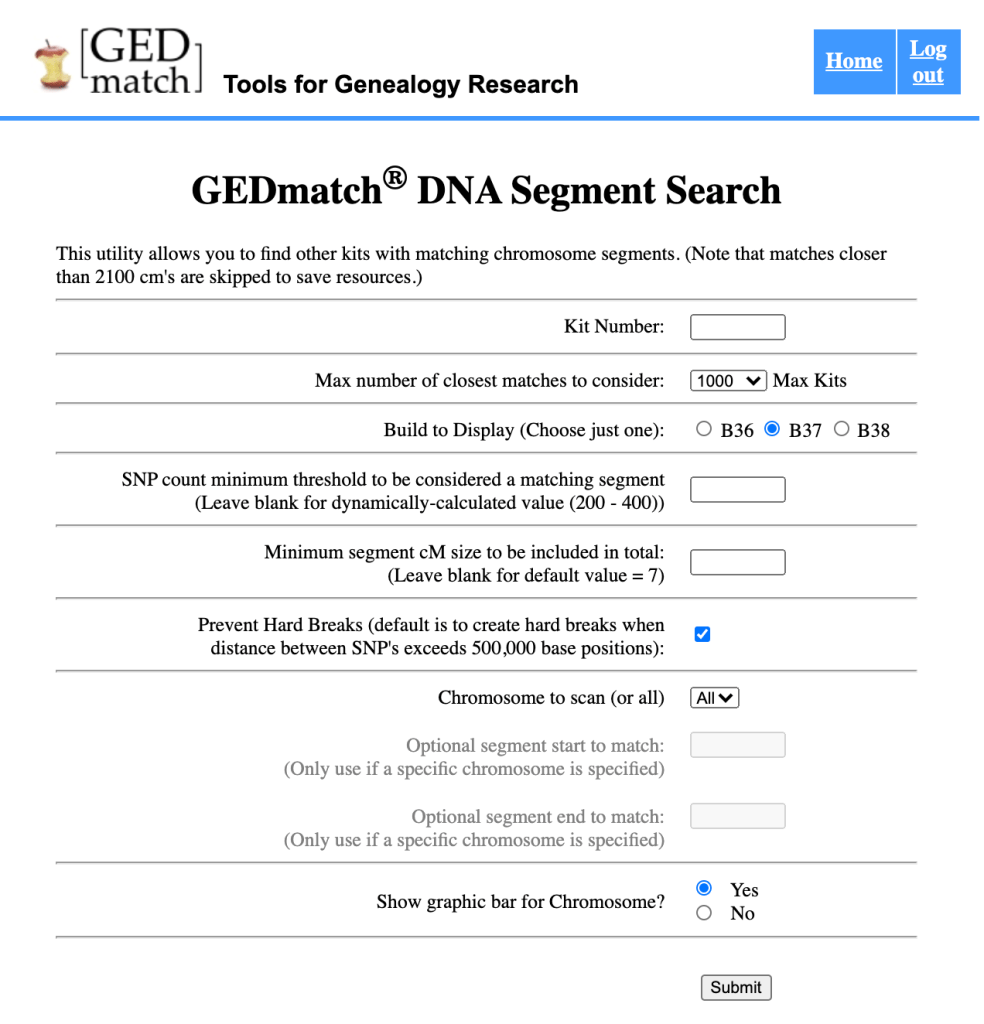
After running your Matching Segment Data you would want to download the csv file. There is a ‘HERE’ button at the top of the list of segment data that allows you to save the csv file to your computer.
Figure 2 shows the GEDmatch Segment Triangulation Screen. It defaults to 500 kits, which I changed to 1000 to match what I’d run on the Segment data. The upper threshold of 3000 cM would exclude parent-child relationships but probably won’t exclude siblings. I left all the other defaults as they were.
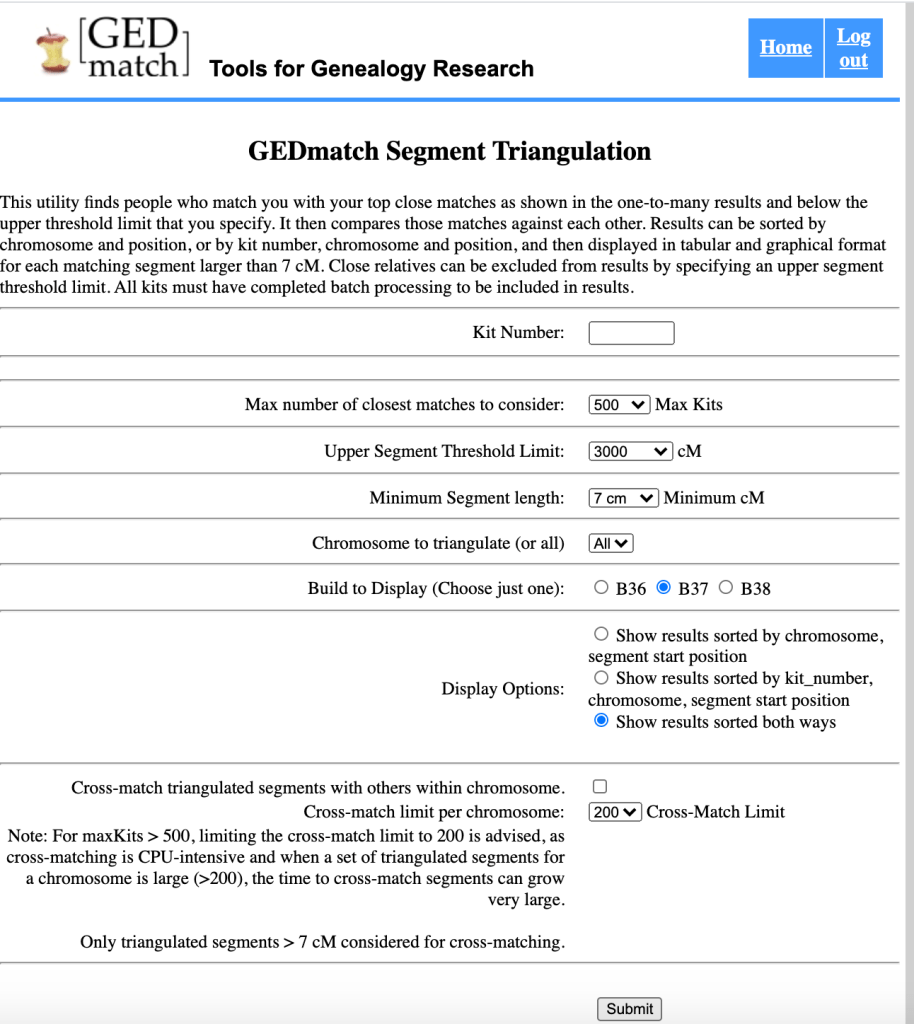
The Segment Triangulation Data can be saved as a tsv file. The ‘HERE’ button to save this data is found at the bottom of the table of triangulated data.
Segment and Triangulation Files
The data in the Segment file shows a list of my matches, the chromosome where we match, how many cM we share, the SNP value, and the start and end of the data on the chromosome. Figure 3 shows the data that I share with a match, Joe1. We share 14.0 cM on chr 6 from about 162 M to 168 M.

When I look at Joe’s triangulated matches in my triangulation file I find that he has 4 matches on chr 6. These data, shown in figure 4, show how many cM Joe, another match and I triangulate in that particular region. It looks as if Joe, John and I triangulate across the entire region that Joe and I match. Whereas the triangulated region for Joe, and Mary or Sue and me is less than the 14.0 cM we share. The data shown in the triangulated data file is showing only the start, end and cM that the three of us who triangulate share. To see how many cM I share with Mary, or John, or Bill or Sue, I’d have to look at the All segment data file. That is why both of these files are needed for the analysis.

With triangulation I’m looking for at least three independent matches that each match me and also match each other. For example a parent and child, or 2 siblings would definitely have a common ancestor, but they would not be independent of each other. The child got half of his or her DNA from that parent, and siblings would share a great deal of DNA in common as well. When the matches triangulate it’s very likely that we share a common ancestor in the genealogical timeframe. Then the next step would be to use traditional genealogy methods to attempt to identify that common ancestor.
Sometimes there are segments found in the triangulation file that are not in the segment file. I downloaded 1000 segment matches and 1000 triangulated matches. Not all segments are going to have triangulated matches, and those segments that don’t have any triangulation will not be included in the cluster. Consequently, since I used 1000 matches for each of the files, there will be segments that triangulate that are not found in the segment file. New DNA matches based on these triangulated segments are reconstructed and these segments are used to estimate the total cM.
Results
Now you are ready to run the GEDmatch enhanced AutoSegment Cluster. Figure 5 shows the data entry page for analysis. Select the maximum cM value you want to include. I pick this value based on what my highest match is and whether or not I want to include that match in the run. The minimum is a bit harder to pick. I know that I don’t have many close cousins at all so I usually pick a low minimum, perhaps lower than most people would choose.
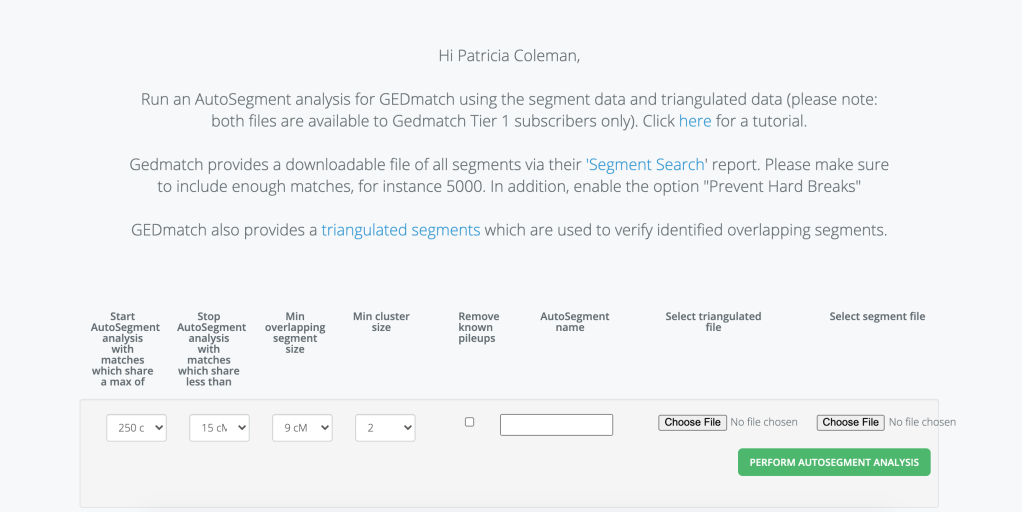
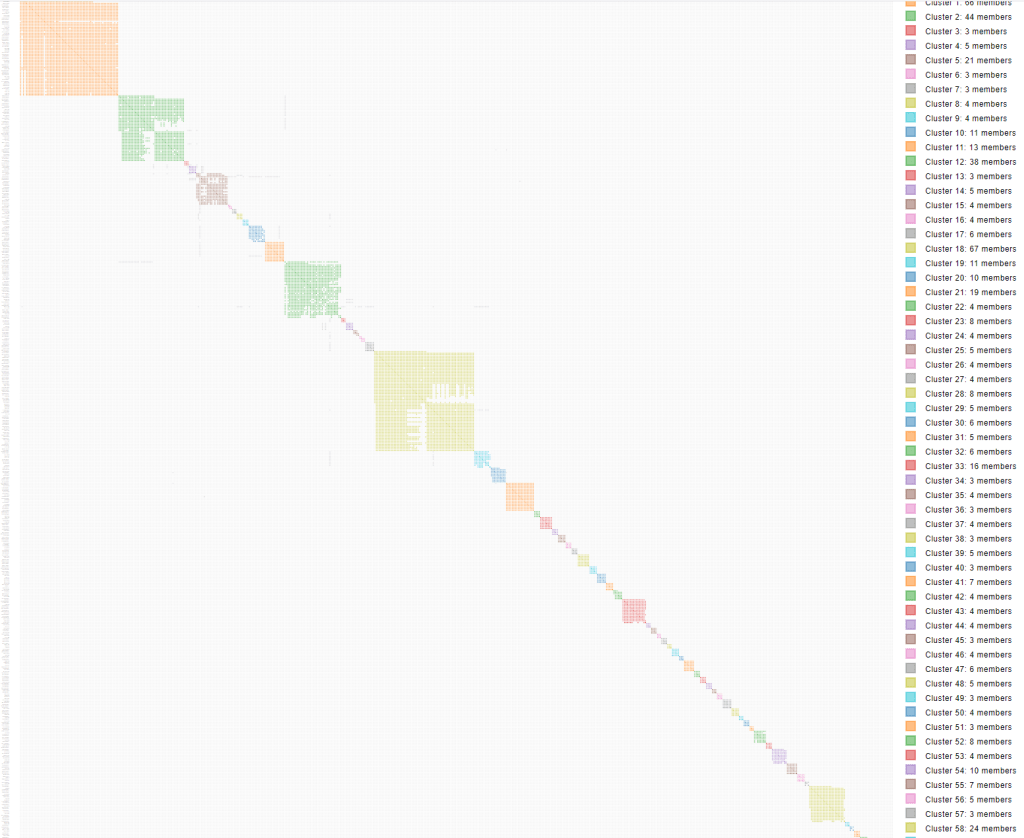
A zip file with the results is sent to your email. My results are shown in figure 6.
Below the clusters are three tables containing information about the clusters and the matches. The first table, shown in figure 7, is the segment statistics for each of the AutoSegment clusters. It describes the segment clusters that are found in each cluster, lists the chromosomes that are present, the number of matches in the cluster, the number of segment clusters and the number of segments. By clicking on the link another window opens showing the segment clusters that make up the large cluster shown in the original html cluster. An example of this is shown below in the Data Analysis.
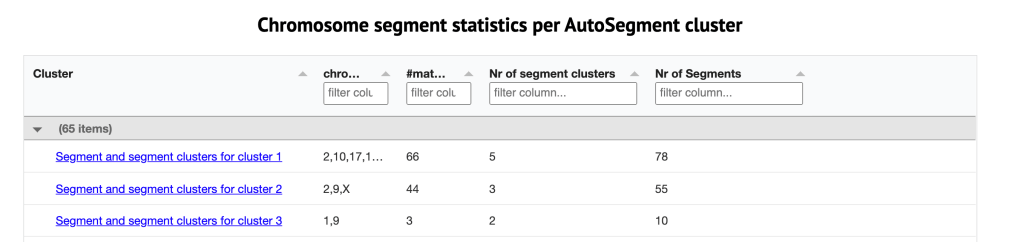
Below this table is the AutoSegment cluster information, which is shown in figure 8. It shows the name and kit number of the match, the amount of cM shared, the number of shared matches, the cluster that this kit in in and other information about the match. The notes indicate the source of the particular segment. Some of this information, such as MyHeritage, and Migration-V4-M, comes from GEDmatch. When the (triangulated) segment is not found in the GEDmatch segment file, Genetic Affairs reconstructs it based on segments that triangulate with it.
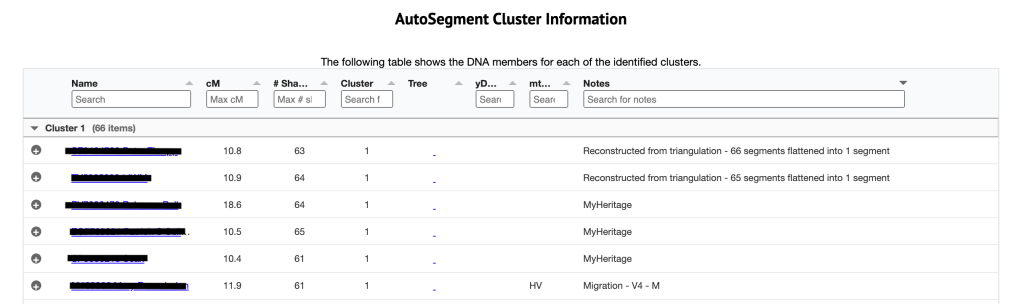
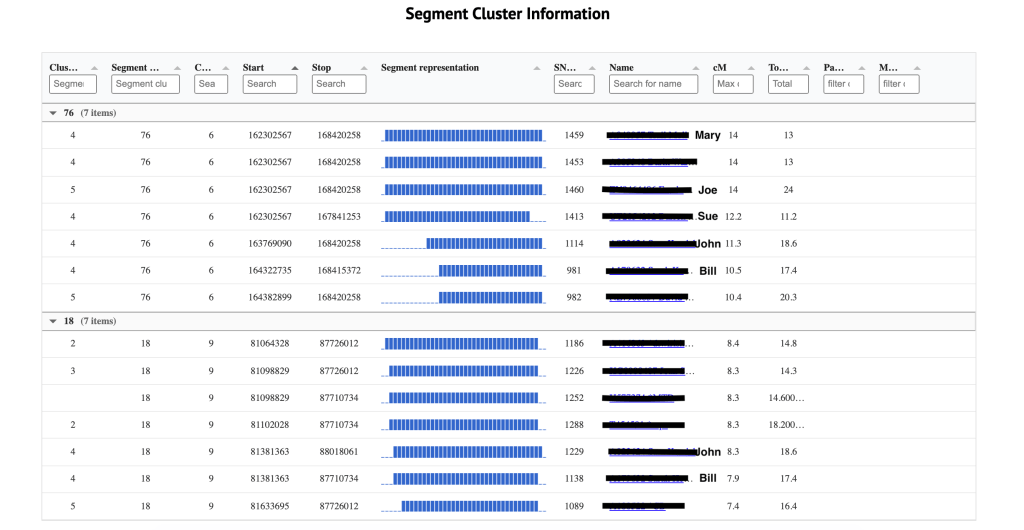
Shown in figure 9 is the third table which is the Individual segment cluster information. The cluster listed on the far left is the cluster number from the large html cluster. Clicking on that number takes me to the segment clusters that are making up the cluster 29. This is the same as if I clicked on the ‘segment and segment clusters for cluster 29’ in the Chromosome segment statistics. Next is the segment cluster number, the chromosome, the start and end values, the SNP, match name and kit, cM for this segment and the total cM for this match. The segment representation chart allows me to quickly assess the overlap between the different segments within a segment cluster.
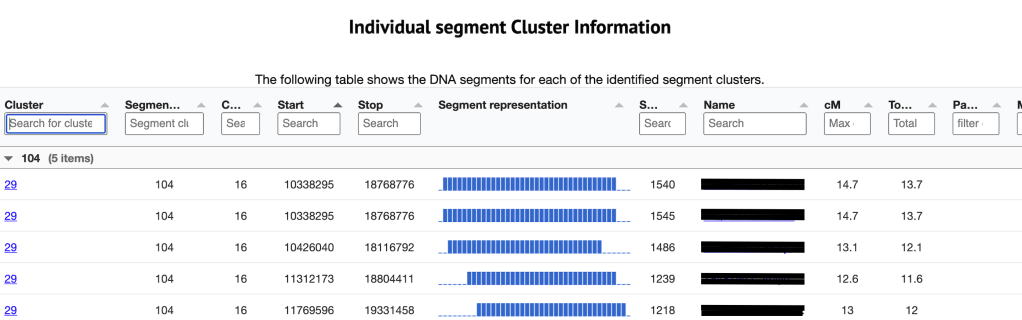
Data Analysis
Earlier I looked at Joe and his triangulated matches on chr 6 in the triangulated segment table. Searching for Joe in the html cluster I found him in cluster 5, the larger brown cluster in figure 6, with grey squares to cluster 4, the purple cluster. Joe matches my known 2nd cousin Frank on chr 20 and was placed into cluster 5 with Frank. You can see the line of grey squares below where I labeled Frank, that are Joe’s matches to John, Bill, Mary and Sue on chr 6 in the purple cluster.
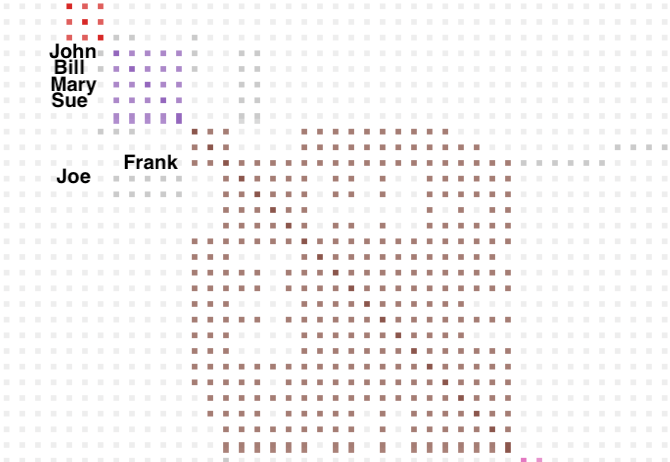
Because of the grey cells Joe will show up in the segment clusters for clusters 4 and 5. Clicking on the link for cluster 4 in the statistics I get another chart with the well known animations which represents the underlying segment clusters and their segment members. This chart allows you to quickly see which and how many segment clusters are present and how connected they are.
For AutoSegment cluster 4, I see there are two segment clusters (blue and orange cluster) in the chart. The orange cluster in figure 11 is the group that triangulates on chr 6. John and Bill also triangulate with another group of matches on chr 9, and those are in the blue cluster.
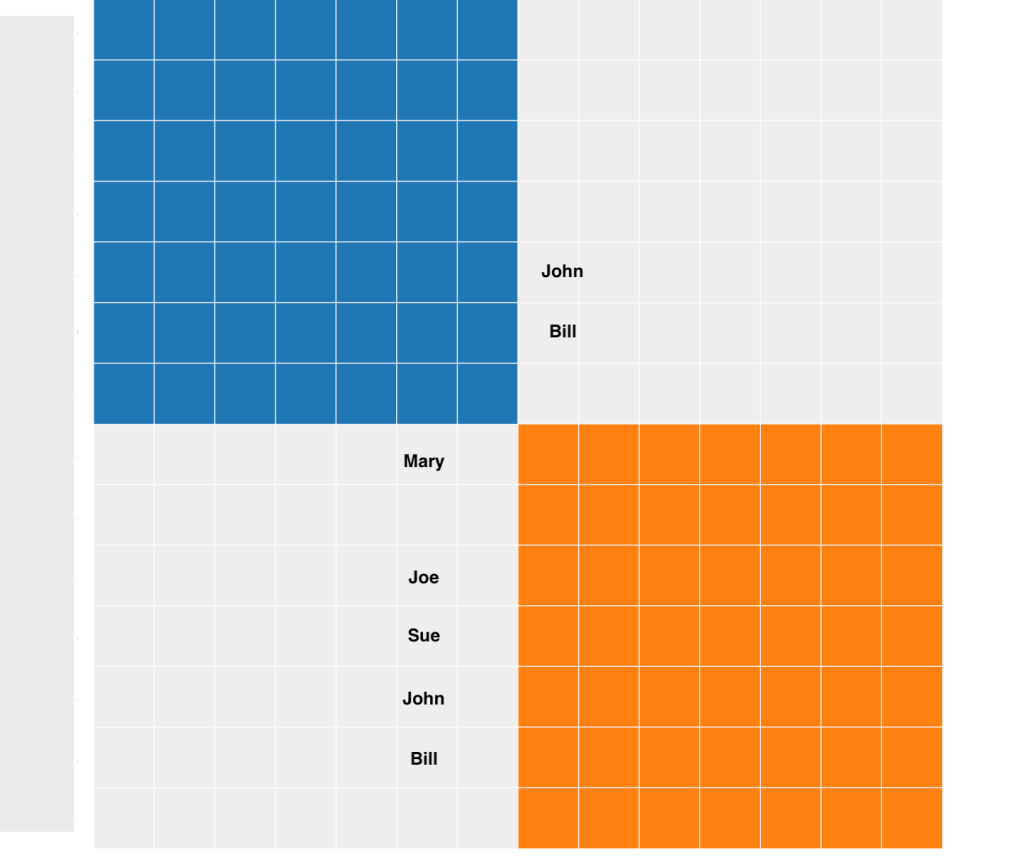
Below these clusters is the Segment cluster information table, shown in figure 12. The column on the left indicates the html cluster where each person was found. The second column indicates the segment cluster. Chromosome 6 has Joe and his triangulated matches. As seen in the clusters John and Bill also triangulate on chromosome 9 with other matches. The start and end values are given and a visual representation of the relative size of the segments are given. The number of cM for each match as well as their total cM are also shown in the chart. The table allows for a quick check how similar the segments are and how they align.
DNA Painter Cluster Tool
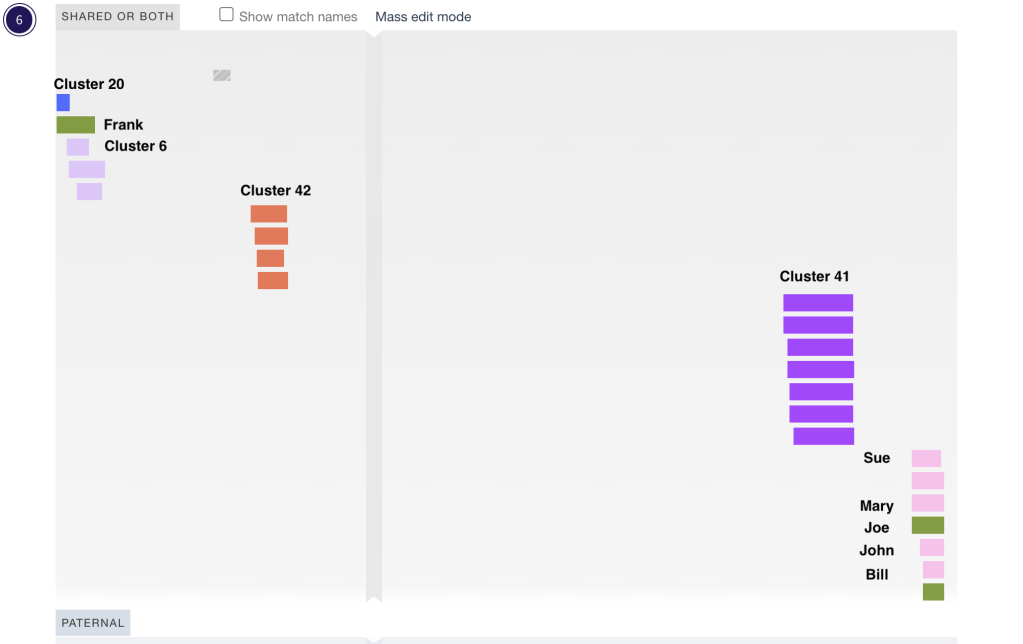
One thing I like to do with my cluster results is to put them into DNA Painter. Using the ‘Cluster Auto Painter’ in the DNA Painter tools I can enter the html file from Genetic Affairs and generate a new profile with all my clusters in it. Figure 13 shows my chr 6 on DNA Painter after importing the html file. Cluster 4 is the pink one on the far right. Joe is on cluster 5 so he’s in a different color. The other green segment in that location is Joe’s son. He would not have been an independent match which is why I left him out of the earlier triangulation. The other 4 triangulated matches are in the pink cluster. I did not add paternal or maternal to any of the matches in my all segment file, so all of my clusters here are showing up as ‘shared or both’. Another thing I’ve done with this DNA Painter profile is to import the GEDmatch segment data file and compare the segments to those in the cluster. It is then very easy to see segments that don’t have any triangulated matches.
Just like the other AutoSegment analyses the GEDmatch AutoSegment clusters costs 50 credits per run.
Summary
The enhanced AutoSegment Clustering for GEDmatch uses the all segment file and triangulation data files from GEDmatch and clusters the matches into triangulated groups. Triangulated groups, especially of four or more, indicate a common ancestor in a genealogical timeframe. Compared to the AutoSegment implementation of MyHeritage, FTDNA and 23andme the GEDmatch version frees users of the manual process of checking the validity of the identified segment clusters.
Individual clusters can then be analyzed using traditional genealogical methods to find a common ancestor. The html file containing the AutoSegment clusters can also be imported into DNA Painter using the ‘Cluster Auto Painter’ tool and visualized in detail on individual chromosomes as well. This is going to be a huge help to me as I research which of our common ancestor my 2C and I share on a particular chromosome segment.
- All names of living individuals have been changed to protect their privacy.
Leave a reply to Paul Baltzer Cancel reply