There’s a new feature at Genetic Affairs – AutoSegment Analysis. Unlike In Common With (ICW) analysis AutoSegment looks at the DNA segments for the analysis. ICW looks at your match list and forms clusters based on shared matches and groups these matches based on them matching the others in the group. The ICW analysis doesn’t look at the DNA or which chromosome contains that DNA, which is why it works for sites that do not have a chromosome browser. AutoSegment first looks at the segments of DNA and where they are located on the chromosomes. Then it groups them based on matches sharing the same segment on the same chromosome (using an user-defined minimum cM overlap) and uses that as the basis for the clustering. This technique uses the downloadable files from the company’s site for the analysis. These are flat segment files without indication as to whether the segment is maternal or paternal for most cases, and you will need to determine that for yourself after you have your AutoSegment results. FTDNA files indicate maternal or paternal if you have placed that DNA match in your tree on the site. Also 23andMe will indicate maternal or paternal if one or both of your parents have tested, or now they are indicating it where they have placed a DNA match in the tree that they are generating for you. Otherwise Genetic Affairs doesn’t know if the data is maternal or paternal. However, after running an AutoSegment cluster and analyzing your results you can edit the original file that you downloaded from the company and add maternal or paternal labels for any match in the list. Then rerunning AutoSegment will indicate P for paternal or M for maternal on the data in the clusters.
Why would you want to sort your data by DNA segments? Recently my 2nd cousin Patricia Ann Harris Anthony, Trish 1, tested on 23andMe. I’ve known Trish all my life. Every summer when I was a kid we’d go to Norfolk, Virginia, where my Dad grew up and spend a week at the beach and visiting family. All of Dad’s mother’s side of the family lived near by. Trish, her mother and grandmother would come down to the beach for a day and visit with us. I was excited to have Trish test. Using her results and mine will help to try and extend that part of our family tree. Our most recent common ancestors are our great grandparents, Thomas Byrnes and Bridget Fenton. Thomas was born about 1840 somewhere in County Roscommon, Ireland, but we don’t know exactly where. Bridget was born in Bruff, County Limerick in 1853. Trish and I share 19 segments of DNA across 14 different chromosome. What I’ve been doing is looking at the matches on these segments, looking at any information on surnames and/or locations they share and building out Quick & Dirty (Q&D) trees to determine which common ancestor of Trish and mine gave us that DNA.
One of the newest tools to help analyze DNA results in Genetic Affairs AutoSegments, shown in figure 1. Data from MyHeritage, FamilyTreeDNA, 23andMe, and GEDmatch can be used for the analysis. Each analysis starts with the data files that are available to download from the company site. The GEDmatch segment data file is available only on Tier1 of GEDmatch.
Since Trish’s data is on 23andMe I started by downloading that data file which is found at the bottom of the page of ‘View All DNA Relatives’ as shown in figure 2.
After selecting 23andMe for analysis in Genetic Affairs the screen shown in figure 3 appears.
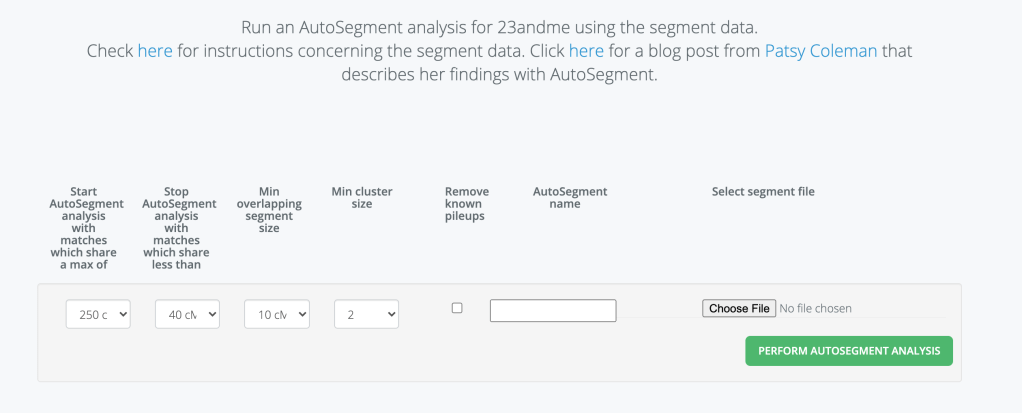
I need to select the maximum and minimum cM values, as well as the minimum overlap cM value for segments. Plus I need to decide if I want to have known pileup regions removed. There are certain regions on chromosomes that have been defined over the years as pileup regions. Typically you might have a large number (over 100) matches in this region which could pretty much be of any size, not just the 7-10 cM variety. Genetic Affairs has added a feature in the AutoSegment analysis to remove those if you chose to do so. Genetic Affairs is using the list published by Li et al (2014)2 for this purpose. The most likely scenario that I can see is that you’d run your AutoSegment Cluster and see some huge number of matches in a particular location. Then check to see if that is a known pileup region that you might want to have removed in a subsequent run. My experience is not finding pileups in the known locations but having what is considered as personal pileup regions. There would be no easy way for software to deal with these. But as I said they are an extremely large region of matches in a specific location. I have around 200 matches on chr 2 in a specific location. When it was only a few I emailed with them and found that they lived in Northern Ireland or their ancestors were from counties in the northern part of Ireland. Since my Thomas Byrnes was from Roscommon this could be likely. I don’t know exactly where he lived in Roscommon and Counties Sligo, Leitrim and Fermanagh are all in the north of Ireland and near Roscommon so it could be possible. When your family is a small and distant as mine you don’t rule out anything that could be possible! But in any case if you find large number of matches in a region that is a known pileup region it might be useful to run the AutoSegment analysis with the pileup regions removed.
A typical range for an AutoCluster analysis is 400 to 50 cM. However, since I wanted to include Trish’s data in the analysis I looked at her match to me to see how much DNA was shared. 23andMe says we share 402 cM, as shown in figure 4.

Therefore I want to set the maximum cM value on Genetic Affairs to the next higher value above 402 cM, which is 600 cM. Setting the lower cM value to some extent depends on your family. My closest cousins are 2nd cousins and so I tend to set a lower minimum value than many people. I’m going to use 30 cM for my minimum. Most people would likely get good results with the minimum set at 40 or 50 cM. My results arrived by email very quickly. I saved the zip file and unzipped it to get the html and Excel files. Figure 5 shows the html file.
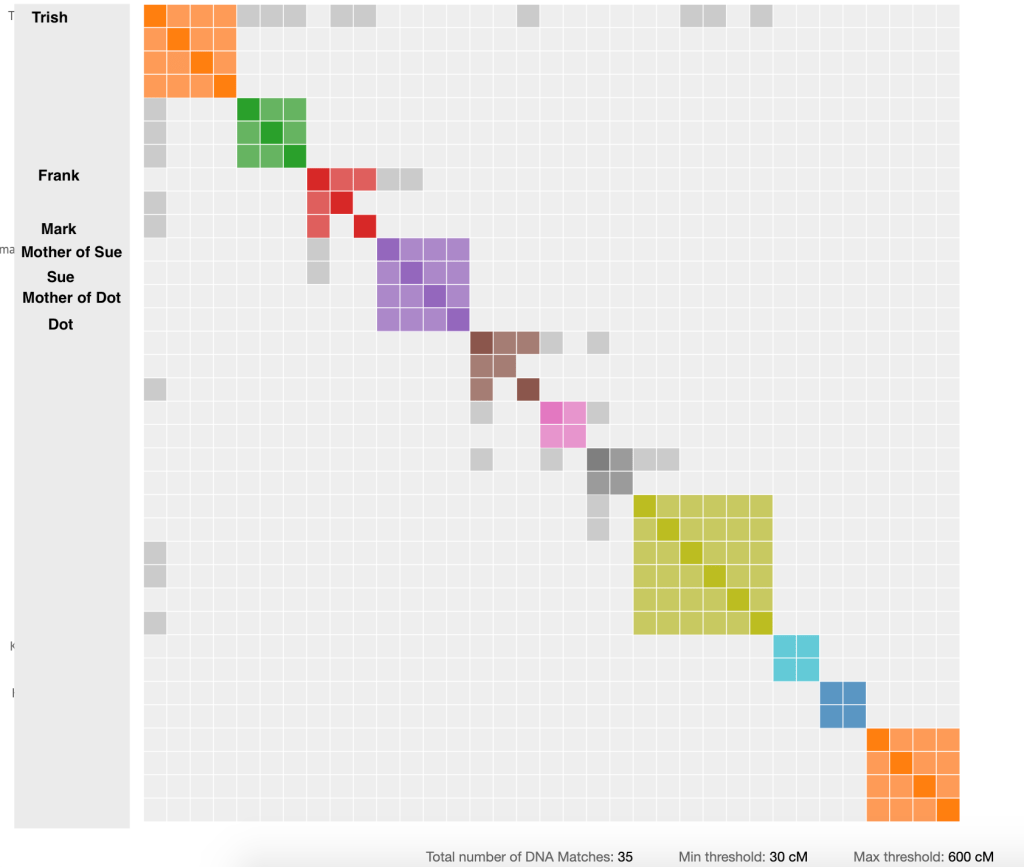
My cousin Trish is in the top orange cluster and matches people in several other clusters. This is not surprising since she and I are 2nd cousins. Frank, who is in the third (red) cluster is also a 2nd cousin. He is on my Dad’s father’s side. I only met Frank six years ago when I started a tree on Ancestry, whereas I’d know my Dad’s mother’s side cousins all my life. I knew my Dad’s father’s side of the family lived in upper state NY. The tree my Dad had done years earlier went through his first cousins, but it didn’t cover their children. But once I started entering the tree from Dad’s notes in Ancestry I found Frank’s tree also had my great grandparents in it, and I sent him a message. Frank and I finally got to meet in person in 2015. Mark is an interesting match since he is shown as being related to both Trish and Frank. Frank also has grey cells connecting him to the purple cluster that has Sue and her mother, as well as Dot and her mother, in it. We’ll investigate these matches further.
At the bottom of the html clusters is a table that explains/expands these clusters. The table is shown in figure 6.

These DNA match clusters are created based on the segment clusters. So someone like my cousin Trish, who shares many segments with me, will appear in a number of clusters. This table shows the various clusters and lists on which of the chromosomes segment clusters are identified. Clicking on any of the cluster links will bring up that particular cluster page. This page will then display the different segments underlying the cluster and the segment clusters in which these segments reside. After looking through the clusters I decided to explore cluster 3 from the list. Cluster 3 is shown in figure 7.

I’ve labeled the matches that were also seen in the AutoSegment html file image shown in figure 5. Cluster 3 is the red one in the html cluster. Because Sue and her mother match Frank and are in cluster 4, Dot and her mother in cluster 4 are included here as well, even though they are not indicated as matching Frank. At the bottom of this html page is a summary of the segment clusters information, which is shown in figure 8.

The most interesting match here is Mark who is shown in the dark blue cluster matching Fred, and in the red cluster matching Trish. Looking at the chromosome segment chart at the bottom of the html page (figure 8) it shows Mark and Fred match on chr 20 and Mark and Trish match on chr 12. From emailing with Mark I know that he and his father triangulate with Fred and me on chr 20 which would be a Barry match, and that our triangulated match on chr 12 is with Mark and his son. I assumed that the chr 12 match was also a Barry match, until Trish’s DNA results arrived. Contacting Mark again I found out that his mother was a Byrnes! This points out how initial assumptions that all the segments of a match are from one most recent common ancestor may not be true. it is definitely a good starting point, but it also needs to be reevaluated when new matches arrive. This is an excellent example of how valuable the AutoSegment tool is to this type of analysis. It also points out how important it is to make contact with matches and to keep chatting with them. Mark and his father match on chr 20 with my Dad’s father’s side, and Mark’s and his son’s matches on chr 12 matches are from his mother’s side and match with my Dad’s mother’s side.
Looking at other matches on this cluster Joe appears to match both Frank and Trish. The only information on Joe’s 23andMe profile is that his paternal grandparents came from Italy. I have no known Italian ancestors, so that was puzzling, and he did not respond to any messages. Using the ‘Advanced DNA Comparison’ on 23andMe shows that Joe doesn’t match either Frank or Trish. Looking at the table in figure 8 it appears that there is plenty of overlap for a match to show up, so that makes me think that Joe is likely on my Mother’s side of the family.
Other interesting matches are Sue and her mother and Dot and her mother. I looked at each in 23andMe Shared matches and found their information. Again I can check them in the ‘Advanced DNA Comparison’ to see if they truly match Frank and/or Trish. But looking at the table in figure 8 there appears to be a very small overlap between Frank and Dot or her mother. Also recall that Dot and her mother did not have grey cells to Frank in the original AutoSegment cluster (see figure 5). Whereas there is a large overlap between Sue and her mother and Frank. The ‘Advanced DNA Comparison’ gives the same results as shown in figure 9. Sue and her mother triangulate with Frank and me and therefore must be paternal on my Barry side.

Their information on 23andMe indicates that Sue’s mother’s maternal grandmother was from the Baden-Württemberg area. My great grandmother Pauline Fröhlich who married Edward Barry was from the Baden-Württemberg area, so that segment on chr 3 is likely from my great grandmother Pauline. Looking at Dot and her mother along with Frank on the 23andMe ‘Advanced DNA Comparison’ shows no match at all. Dot and her mother could be maternal matches or perhaps there’s not enough overlap to Frank to show as a paternal match. I’ll leave them as ‘unknown’ for now.
This is only the analysis for one of the cluster links from the table, shown in figure 6, that was located at the bottom of the AutoSegment cluster html file. All of the links in that table could be explored for similar information and new findings.
Another new feature for AutoSegment is being able to add maternal or paternal to the original data file. My file was downloaded from 23andMe. Opening that csv file in Excel allows me to add maternal or paternal in a new column at the end of the existing file. The 23andMe file is in the form of name_relatives_download.csv For this file the last filled column is AD so I’ll be added maternal or paternal in column AE. I search column A for my cousin’s name – Trish Harris Anthony. I found her starting in row 648 as shown in figure 10.

Next I’ll scroll to the end of the row and place paternal in column AE for all the rows in which she is listed, as shown in figure 11.

I’ll do this for Frank and other matches that I know where they fit in my family. Then rerunning Genetic Affairs AutoSegment will include that information in the html file display, as shown in figure 12. Besides adding Trish and Frank as paternal I added maternal to a couple of known matches in cluster 8. Looking at the html AutoSegment cluster in figure 12 you can see P next to Trish’s name and next to Frank’s name for paternal. Joe and Mark are also considered paternal, since they match both Trish and Frank. I know that Mark is paternal, but having already looked at Joe’s match in the ‘Advanced DNA Comparison’ on 23andMe I know that he doesn’t match either Trish or Frank. His being marked as paternal is a hint, just like any other hint you get from looking at DNA results and needs to be checked out, which I’ve already done.

Another thing that can be done with the AutoSegment html cluster is that it can be added to DNA Painter using the ‘Cluster Auto Painter’ found in the DNA Painter tools. I uploaded the html file shown in figure 12. The DNA Painter profile is shown in figure 13.
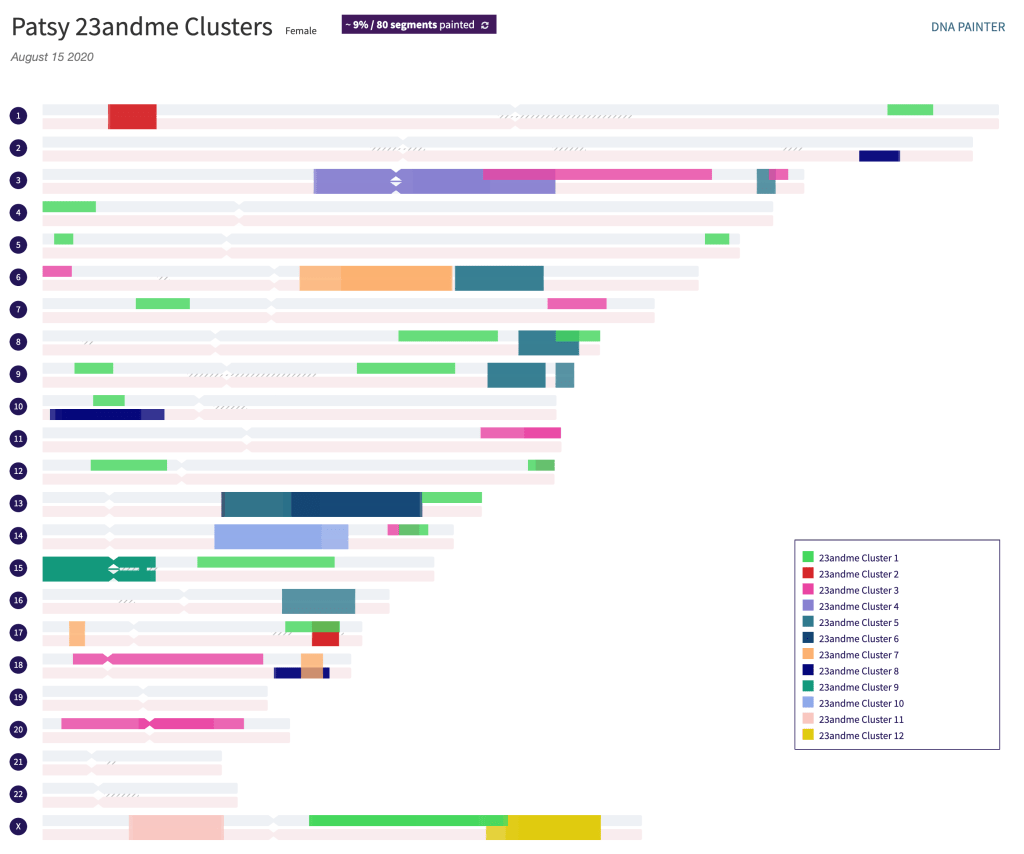
Notice how the light green for cluster 1 and the pink for cluster 3 are shown as paternal, and dark navy for cluster 8 is shown as maternal, while the others are all shown as ‘unknown or both’. This is particularly apparent on chr 10 where you can see the light green paternal and the dark navy maternal segments near the beginning of the chromosome. Figure 14 shows the expansion of cluster 1 with part of the segments where Trish matches me, and all are listed as paternal. One of our shared matches who triangulates with the two of us on the X chromosome also has a segment on chr 5. This might be a useful segment to investigate further for other matches that could potentially also be on my Byrnes and Fenton line.
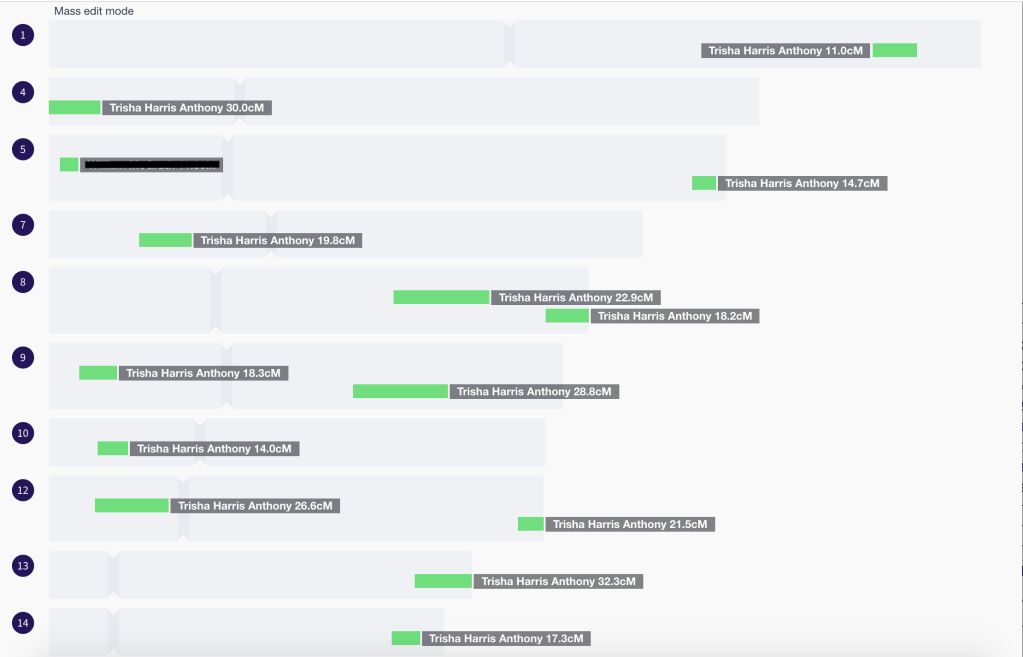
Remember Sue and her mother and Dot and her mother that were in cluster 3 from figure 7 and 8? Now we can look at them in DNA Painter and change them from ‘unknown or both’ to ‘paternal’ for Sue and her mother and ‘maternal’ for Dot and her mother. Figure 15 shows the four of them in cluster 4 as was seen in the html file (see figures 5 and 12).
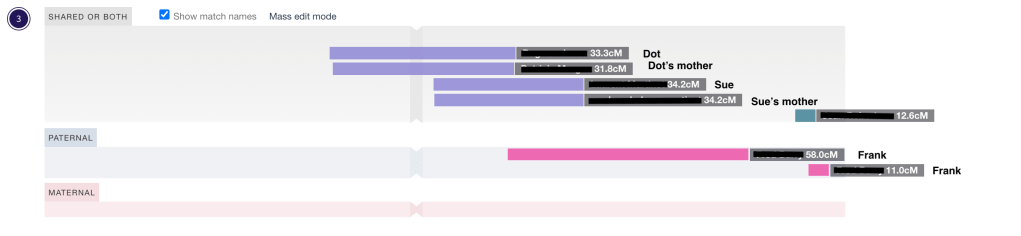
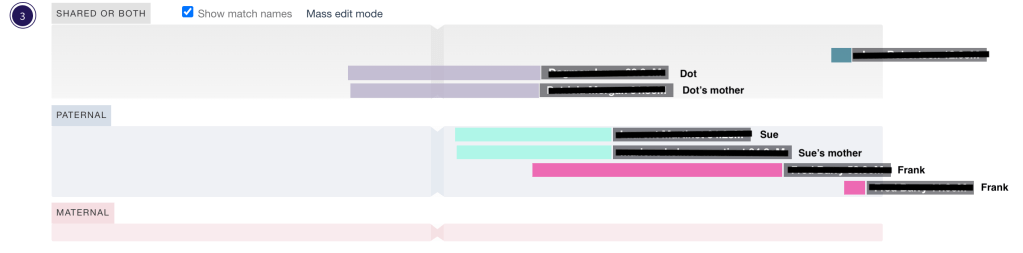
Using DNA Painter’s ‘mass edit mode’ I moved Sue and her mother to ‘paternal’ since they triangulated with Frank on chr 3 when I tested that in 23andMe ‘Advanced DNA Comparison’. I then moved Dot and her mother to ‘maternal’ since 23andMe ‘Advanced DNA Comparison’ showed that they did not match Frank at all. Chromosome 3 after moving Sue and her mother and Dot and her mother is shown in figure 16.
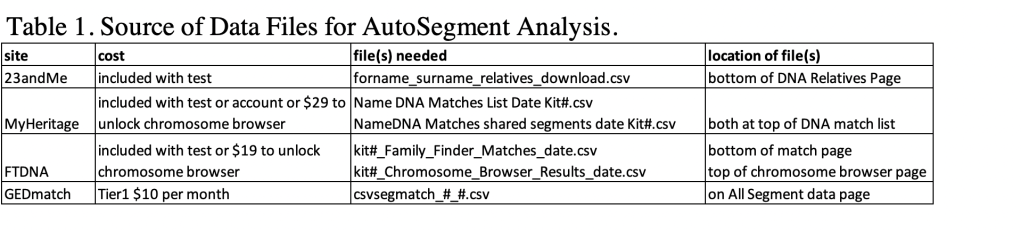
All of the analysis I’ve done here has been with 23andMe data. The same thing can be done with your DNA matches from FTDNA, MyHeritage or GEDmatch data. Table 1 shows what data files are needed, and where they are located. If you tested at FTDNA or MyHeritage the chromosome browser at FTDNA and DNA tools at MyHeritage is included in the price of your test. If you uploaded your data FTDNA charges $19 to unlock the chromosome browser. MyHeritage charges $29 for the DNA tools, unless you unloaded your DNA data and have an account with them. MyHeritage allows a tree up to 250 people for free, but larger trees, such as I have, requires an account. A summary of the location, and filenames for the various sites that can be used for Genetic Affairs AutoSegments analysis are shown in table 1.
On Genetic Affairs website you can run ICW AutoClusters for 23andMe and FTDNA data but not for MyHeritage nor GEDmatch. MyHeritage has an early version of the Genetic Affaires software in their DNA Tools. It does not allow you to select the max and min, and gives you results with about 100 matches in the clusters. GEDmatch also has a version of Genetic Affairs software that allows you to set max and min but uses their unknown algorithm to determine how they use this range. Now with AutoSegment analysis you can run all the analyses on Genetic Affairs website. You can select whatever max, min and minimum shared segment size that you want to use. No longer do you need to give your login credentials in order to run this analysis. That has been a concern for a number of people in the past.
What does the AutoSegment analysis cost? There is a new free tier for AutoSegment. For MyHeritage, 23andMe and GEDmatch an analysis of 250 – 25 cM with a 15 cM segment size is within the free tier. For FTDNA a 250 – 45 cM size with 15 cM segment size is also within the free tier. Those will give you a chance to play with the new feature and potentially get you hooked on it, as it has me! AutoSegment Cluster of larger, different sizes cost the same prices as ICW clusters. Generally they are around 50 credits per run.
In summary I find this new feature very exciting. I was already starting to look at the segment Trish and I shared so this makes that analysis so much easier. The AutoSegment analysis can be run with data that you download from 23andMe, FTDNA, MyHeritage or GEDmatch. You get to select the maximum, minimum and the amount between shared matches. You don’t have to provide your login information to the other sites since you’ve downloaded the files they make available to you from their sites. You can put the resulting html file into DNA Painter using their ‘Cluster Auto Painter’ tool. And you can add maternal or paternal to known matches in the data files from the companies to better sort your matches.
- Patricia Ann Harris Anthony has given me permission to use her real name. All the other names used in this post are fictitious.
- Li et al., “Relationship Estimation from Whole-Genome Sequence Data”, Plos Genetics, 10, (Jan. 2014), 1371.
Leave a comment