An exciting addition to Genetic Affairs is Hybrid AutoSegment Clusters! Now you can run the AutoSegment clusters with data from 23andMe, FTDNA, MyHeritage and GEDmatch or any combination of these sites all into one cluster analysis. The entry page is shown in figure 1.
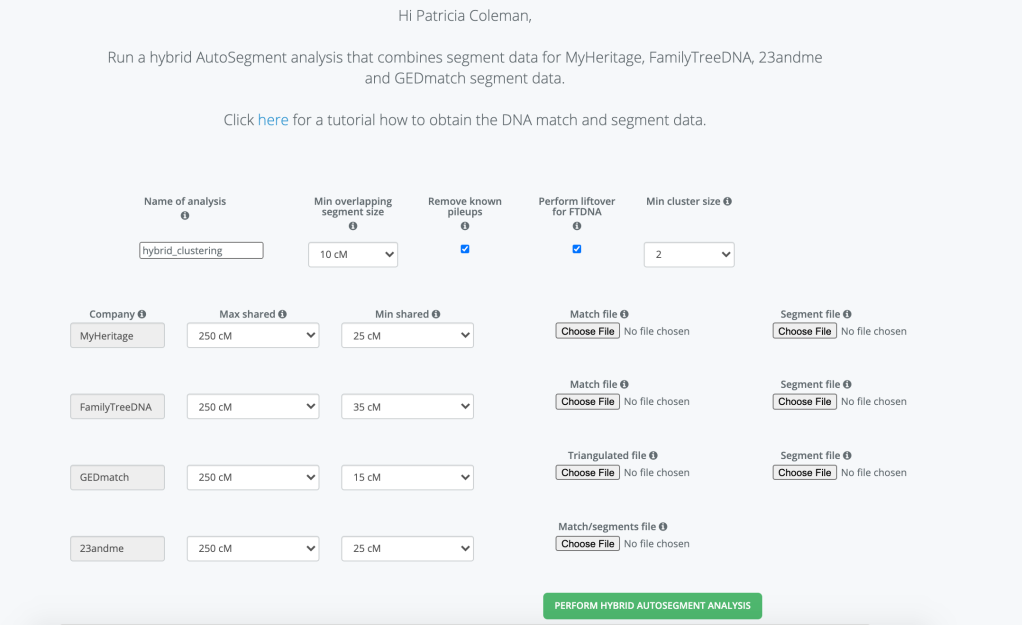
Starting at the top of the page you’d want to give a name for you Hybrid cluster. I often use the name of the person whose cluster it is and a date or some information that will tell me exactly what the file is. You can select the minimum overlapping segment size between your matches, and the minimum cluster size. The smaller the overlap and the smaller the cluster size the more matches that will be used, and it could end up with an html cluster that is too large for your browser to load. You can always view it with the Excel file or look at the html file that has all the information without showing the large cluster.
If you have a large number of matches in known pileup regions you can choose to have those matches removed from the analysis. Pileup regions are explained in more detail at Genetic Affairs
Another parameter to consider is liftover for FTDNA. Of the various testing sites only FTDNA still uses build 36 for their comparison whereas the other sites are using build 37. A build is a reference system used by the testing company that represents the human genome. For comparing matches across the different companies I would want all the data to be using the same reference. Performing liftover on the FTDNA matches converts them to build 37, so that you can easily do a direct comparison.
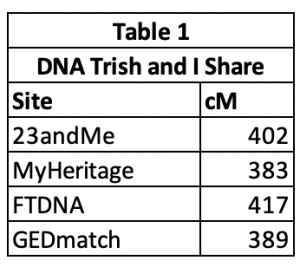
You can select different min and max cM settings for each of the sites. What I typically do is to look at the site and select a max cM value that will include the highest match that I want in the analysis. My paternal 2nd cousin Trish1 tested at 23andMe and uploaded to MyHeritages and GEDmatch. She did mtDNA and Family Finder tests at FTNDA. But each of the sites reports a slightly different cM that she and I share. Table 1 shows the amount of DNA Trish and I share at each site. Both 23andMe and FTDNA include our X chromosome match, as well as FTDNA counting small cM down to 1. I could run MyHeritage and GEDmatch with a max of 400 cM, but if I used 400 cM for all of the sites, I’d not include Trish’s data at 23andMe and FTDNA. I usually just use 600 cM max for all four of the sites, since I know Trish is my highest match it’s not hurting anything to have the max higher than needed.
I find it harder to select a minimum cM value. I’d like to go down to around 7 cM, but then the clusters are so large that it’s very difficult to load and view them, at least on my laptop. Minimum shared is the amount shared between your DNA matches. And minimum cluster size is the number of matches needed to make a cluster.
The match and segment files that are used in the analysis are the ones that you download from the particular site. You can select to run two, three or all four sites. If you want to run just one site AutoSegment you should use the AutoSegment Analysis from Genetic Affairs main page. The cost of the Hybrid AutoSegment Analysis is 100 credits. It is not part of the free tier which is the 200 credits you receive when you first join Genetic Affairs. For the paid tier you can make a one-time purchase of any amount. For example, a $5.00 (USD) would purchase 5.00 credits. Or you can select to have a monthly subscription for as little as $5.00 (USD) per month. Monthly subscriptions also provide 10% additional credits. So a $6.00 (USD) subscription will result in 660 monthly credits.
When your hybrid AutoSegment cluster is ready you will receive an email. If the resulting file is less than 8 MB, the zip file will be attached to the email. For larger files the email will contain a link where you can download your results file.
Results
This is my beautiful html cluster from 600 cM to 25 cM on all 4 sites: MyHeritage, 23andMe, FamilyTreeDNA and GEDmatch, shown in figure 2. The segment clusters from MyHeritage, 23andMe and FTDNA look at segments that overlap on a particular chromosome. In general they are not considering maternal or paternal. FTDNA will label a DNA as maternal or paternal if you have identified a match in your tree. Maternal or paternal is sometimes indicated at 23andMe. You can also add maternal or paternal to known matches in the CSV files after retrieving them from the testing company. The GEDmatch data uses both the triangulated data as well as the segment data, so the results for GEDmatch are triangulated segments. If you know one match in the GEDmatch data in a particular cluster is paternal, you know that all the GEDmatch segments in that cluster also have to be paternal because of the triangulation.

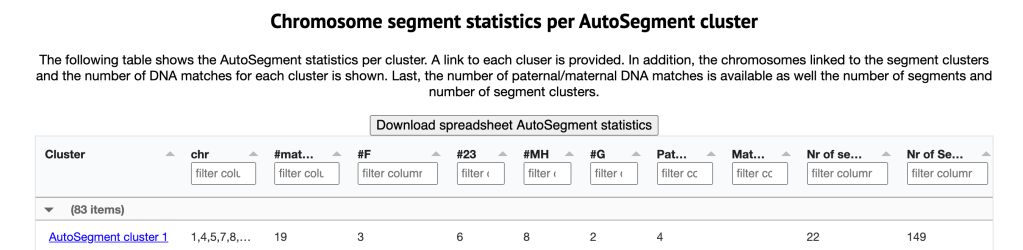
That first orange cluster with lots of grey squares has my paternal 2nd cousin Trish in it. Since Trish is on all four of these sites she’ll show up as four matches. The table below the html cluster contains the chromosome segment statistics per AutoSegment cluster, shown in figure 3. This table contains a link that will bring up a more detailed page concerning the cluster of interest as well as provide some information concerning the identified segment (clusters) such as the chromosomes underlying the segment clusters, how many matches (per DTC) and if there are any maternal or paternal annotations linked to these clusters.
Clicking on the AutoSegment cluster 1 link in the table brings up a visualization of the identified segment clusters and the individual segments that have made up cluster 1 in the html file. These segment clusters are shown in figure 4. A colored square is present between two segments indicates that there is sufficient overlap between those segments.


Figure 5 shows the segment cluster information for segment cluster 13, the red one about in the middle of figure 4.
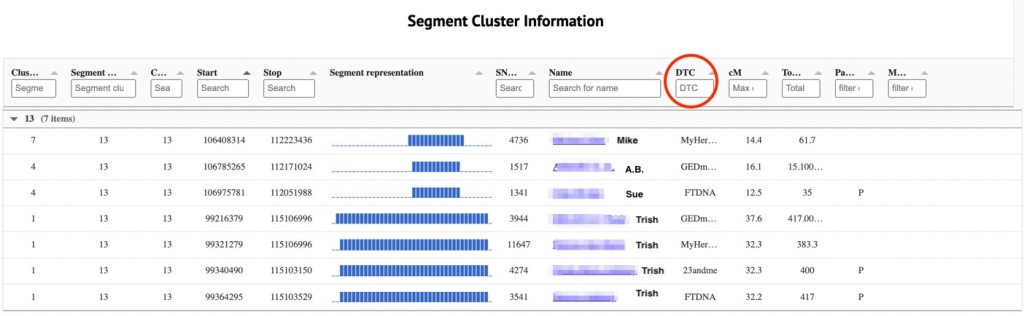
There are 7 matches listed here. The first column tells the cluster number where the match was found in the html cluster. The second column has the segment cluster number, here all are 13. Next is the chromosome number, which happens to be chromosome 13 here. Then the start and end values of the segment. The diagram is a visualization of that segment of data. You can easily see that all 7 of these overlap. The SNP value is in the next column. Followed by the name and kit number of the match. I added a red circle around DTC, DNA Testing Company. The next column has the number of shared cM on this segment, followed by the total number of cM that the match and I share. The last 2 columns have paternal and maternal if that information is found in the file.
The last 4 matches are all of Trish from the 4 sites. Mike tested at MyHeritage. I know that Mike is a maternal 2nd cousin once removed. He and I share my maternal great grandparents as our most recent common ancestor. He has a segment of DNA on chromosome 13 that overlays Trish’s segment. But because MyHeritage is giving all segments that fall in the same location he and Trish show up in this cluster. If I didn’t know who Mike was, I’d go to MyHeritage and run the chromosome browser with Trish and Mike in order to see if they triangulate with me. Next is A.B. whose data came from GEDmatch. Because the GEDmatch segment data here is triangulated I know that A.B. must be paternal because he triangulated with Trish. Sue is from FTDNA and has a P off to the far right, which tells me that I’ve placed her in my tree, and FTDNA knows that she is paternal. If she were totally unknown I’d use the FTDNA chromosome browser and matrix to determine if she matched Trish or not.
Having the data from the different sites displayed this way makes it’s easy to see matches that overlap and might be related. Then you can check in the chromosome browser on the individual testing company site to confirm if they are on the same side of your family or one is maternal and the other is paternal. It’s especially helpful when I find a match that I’ve not looked at before, and now I have some idea how the match might relate to me based on who else is on that DNA segment and in the cluster.
I’ve done a lot of research with matches that my cousin Trish shares with me, so I decided to look at some more distant matches. Searching through the list of names in the Excel file I found Sophie who is in this cluster 70 which I’ve circled in the large html cluster in figure 6.
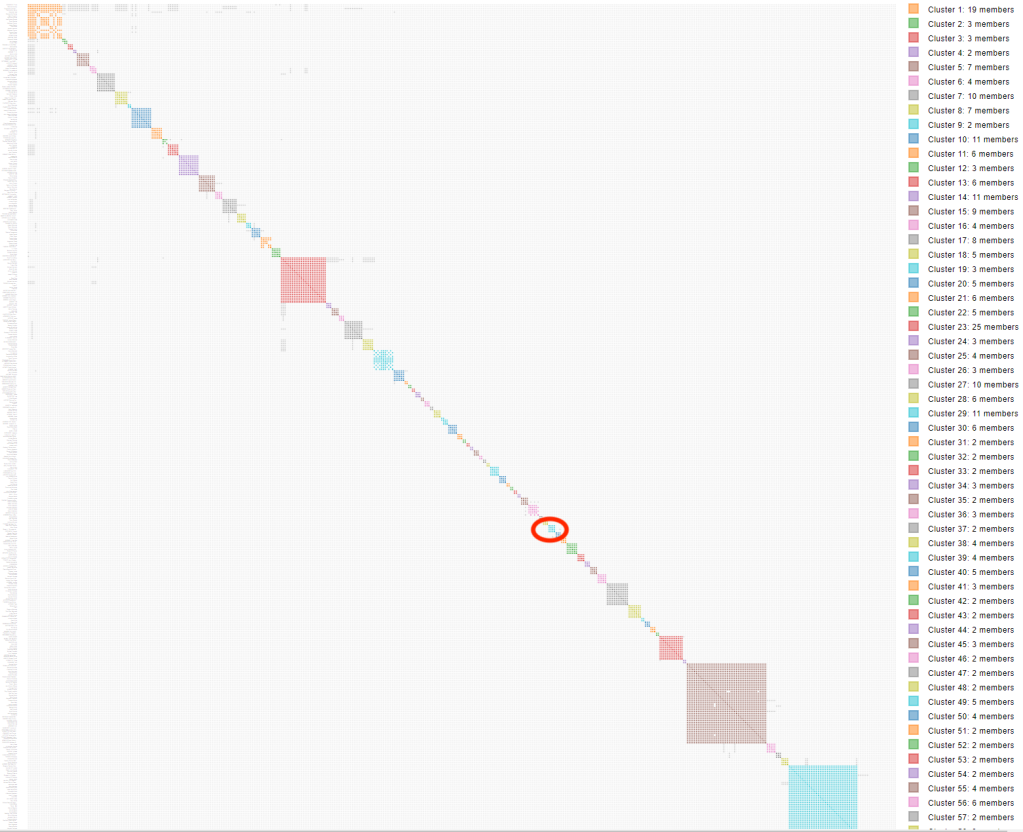
Figure 7 shows the segment clusters chart using a visualization of the individual segment. I have no idea who Andrea is, other than she matches Sophie.
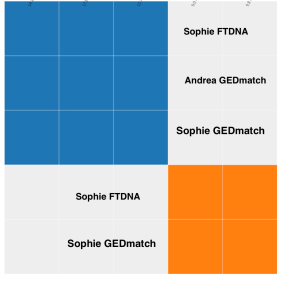
Sophie tested at FTDNA and uploaded her results to GEDMatch. She shows up here matching herself and Andrea. Sophie and I have emailed a number of times. We know that our comment ancestor is on my Aide line. My 2nd great grandfather Thomas Barry married Mary Aide in County Kilkenny, Ireland. I have the baptismal records for their two children, Edward, my great grandfather, and Mary his sister. Edward was baptized in 1840 and Mary in 1843. None of the baptismal records prior to 1823 and none of the marriage records for the Catholic parish in Ballyhale, Kilkenny survived. So I’ve not been able to find Mary Aide’s baptismal record or Thomas and Mary’s marriage record.
Sophie’s Aide family also lived in County Kilkenny, and goes back another generation or two past my Mary Aide. Mary Kilfoil married an Aide, and as best as we can tell without records and with the DNA evidence Mary Kilfoil is either my 3rd or 4th great grandmother. This continues to be something that I’m searching, but for now we’ll leave it at that.
Andrea on GEDmatch indicated that she’d tested at 23andMe. Almost to my surprise I found her in my match list on 23andMe! She had no triangulated matches with me, which was a disappointment as I like to work with triangulated matches. But looking at her ICW match list, shown in figure 8, was amazing! Frank Barry, my 2C who also descends from Thomas Barry and Mary Aide is at the top of the list. Looking down the list I’d already added many of her matches and their triangulated matches to my DNA Painter profile. I’d messaged Tyler over a year ago on 23andMe and never got a reply, so I really don’t have any information on him. He does triangulate with known Irish matches, however. Kay and I have emailed a good bit. She has a great grandfather surname Byrne from County Roscommon. I have my great grandfather, Thomas Byrnes, from County Roscommon. We’ve not found the common ancestor yet, but the connection seems to be on my Byrnes side. Beth is a bit of an unknown as she and Trish have segments on the same chromosome and somewhat overlap, but don’t show as a match. Either Beth is on my maternal side or there’s just not enough overlap with Trish. I need to message her for more information. Ashley was a match I’d not looked at before. So I looked at her shared matches and any information they might have listed. I found one of her matches with ancestors from Buffalo, NY. Thomas Barry’s family lived in Evans, Erie County, NY, which is not far from Buffalo.
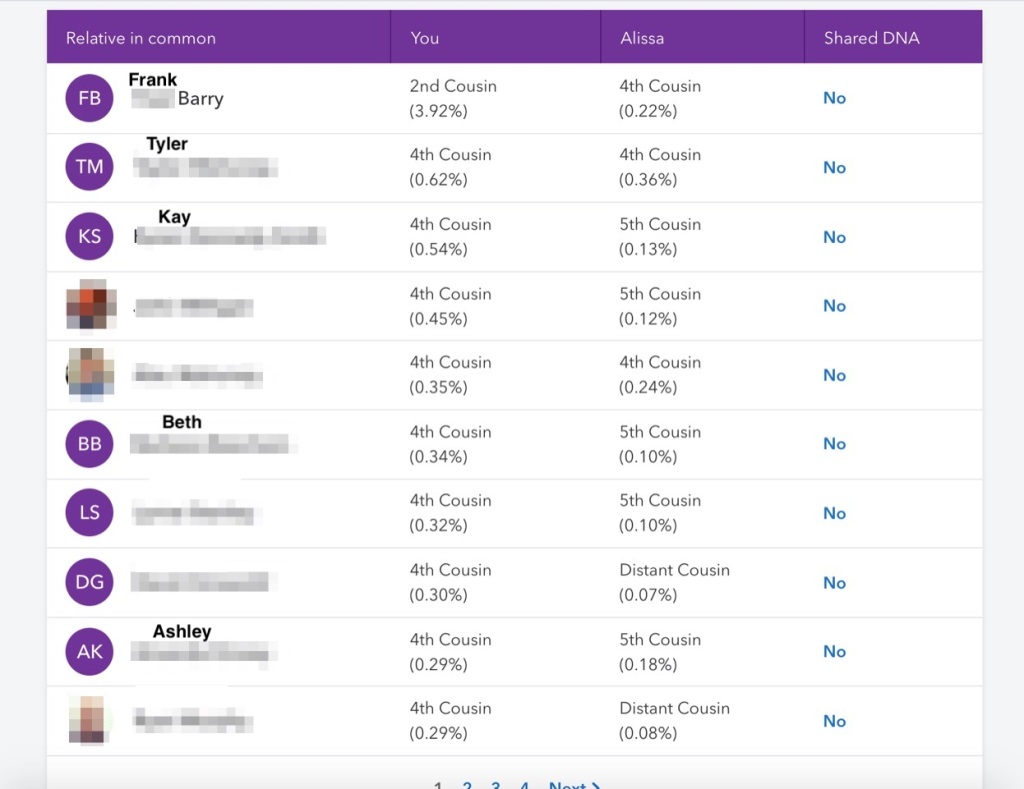
One of the surnames, Green, and one of the locations on Andrea’s information on 23andMe were the same as I knew were in Sophie’s family. And not finding much information from Andrea’s matches I emailed Sophie. Sure enough Andrea is on the same line as Sophie and is Sophie’s 2nd cousin once removed. Andrea is another match on my Aide line. Now if I could just make the connection to our two trees and figure out if Mary Kilkoil in my 3rd or 4th great grandmother! I’ve tried WATO, but most of my matches to Aide family members are too small to be useful in WATO, so I haven’t gotten very promising results.
Summary
I’m finding the Hybrid AutoSegment Clusters on Genetic Affairs very promising. There are so many new connections for me to explore! I would not have found Andrea and been able to connect her to Sophie if not for the hybrid clustering. Sophie has not tested on 23andMe. Andrea didn’t have any triangulated matches there. At most I’d have seen the Green surname, and since it’s not that unusual a name I might not have ever thought of Sophie and that it’s in her family tree. The Hybrid AutoSegment Clusters is going to be a huge help for me trying to make connections between more of my DNA matches.
You can run the AutoSegment Clusters with any 2, 3 or 4 of the testing companies: MyHeritage, 23andMe, FTDNA and GEDmatch. It will provide you with clusters that are based on shared segments across the companies that you selected. With the exception of GEDmatch, where the data has already been triangulated, you will need to compare matches in one of the segment clusters with each other using the chromosome browser, and on FTDNA the matrix tool, to determine if the matches triangulate or not.
Now to go explore more of my hybrid clusters!
- Patricia Ann Harris Anthony, Trish, has given me permission to use her real name. All the other names used in this post are fictitious.
Patricia,
Great write up and example. Now I need to find the time to experiment and play with this.
LikeLike
Thank you for sharing this!
LikeLike
Ideally I would like to generate 8 clusters, one for each great grandparent. Is this resonable? Playing with the thresholds I’ve never been successful at doing this, however. Any ideas? Thank you.
LikeLike
It would all depend on if/how many of the descendants of these great grandparents have tested. AutoSegment is also looking a overlapping segments and not ICW so that would make it more difficult as well.
LikeLike